G-RISE Trainee Program Overview
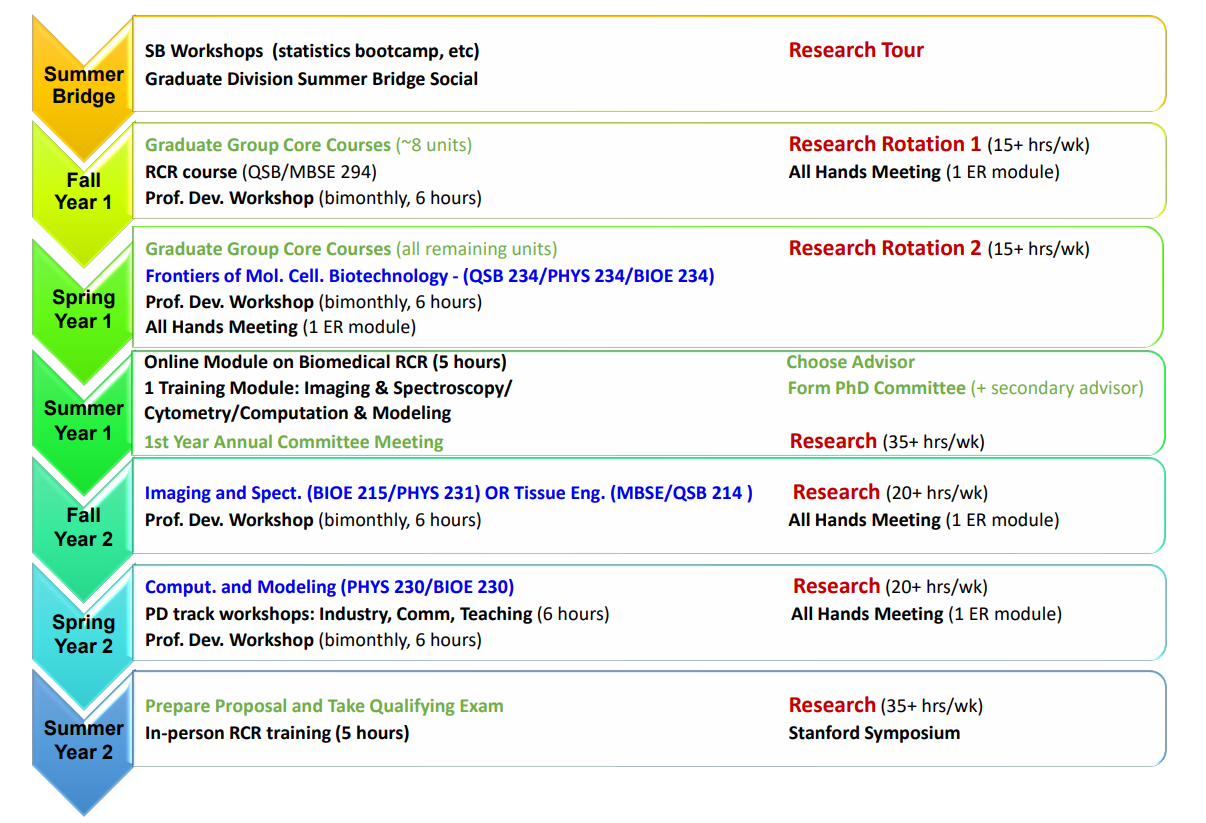
Once admitted to the program, G-RISE students are expected to complete specified coursework and training within the two first years of their Ph.D. career. The program activities are designed to prepare a diverse pool scientists earning a Ph.D. for competitive careers in the biomedical field by training students to identify and solve pressing biologicalproblems using quantitative interdisciplinary approaches
Further details about Coursework
NIH Grant Number: T32 GM141862
Please use this grant number in acknowledgements for posters and publications
G-RISE Trainee Program Timeline